Evolution and the Microbiome
Given the three main goals of evolutionary biology – to discover and describe biodiversity (including estimate phylogeny), to elucidate the natural history and lifestyle of organisms, and to understand the processes that generated the history and phylogeny – it is logical to include the microbiome in evolutionary biology as both a trait worth investigating and as organisms worth understanding. Primarily, we’re interested in how the host organism affects the microbiome and how the microbiome affects the evolution of the host. This is a “Big Question” that will require many incremental steps – e.g., what is the natural variation in the microbiome across populations within a host species?

The Avian Microbiome
Birds exhibit immense variation in their microbiomes – both across the entire class Aves and within single species. Their unique adaptations – flight, feathers, eggs, migration, parental care, etc. – have almost certainly shaped their microbiomes and there is much, much work to be done in understanding how bird life, history and behavior are reflected in or affected by the microbiome.
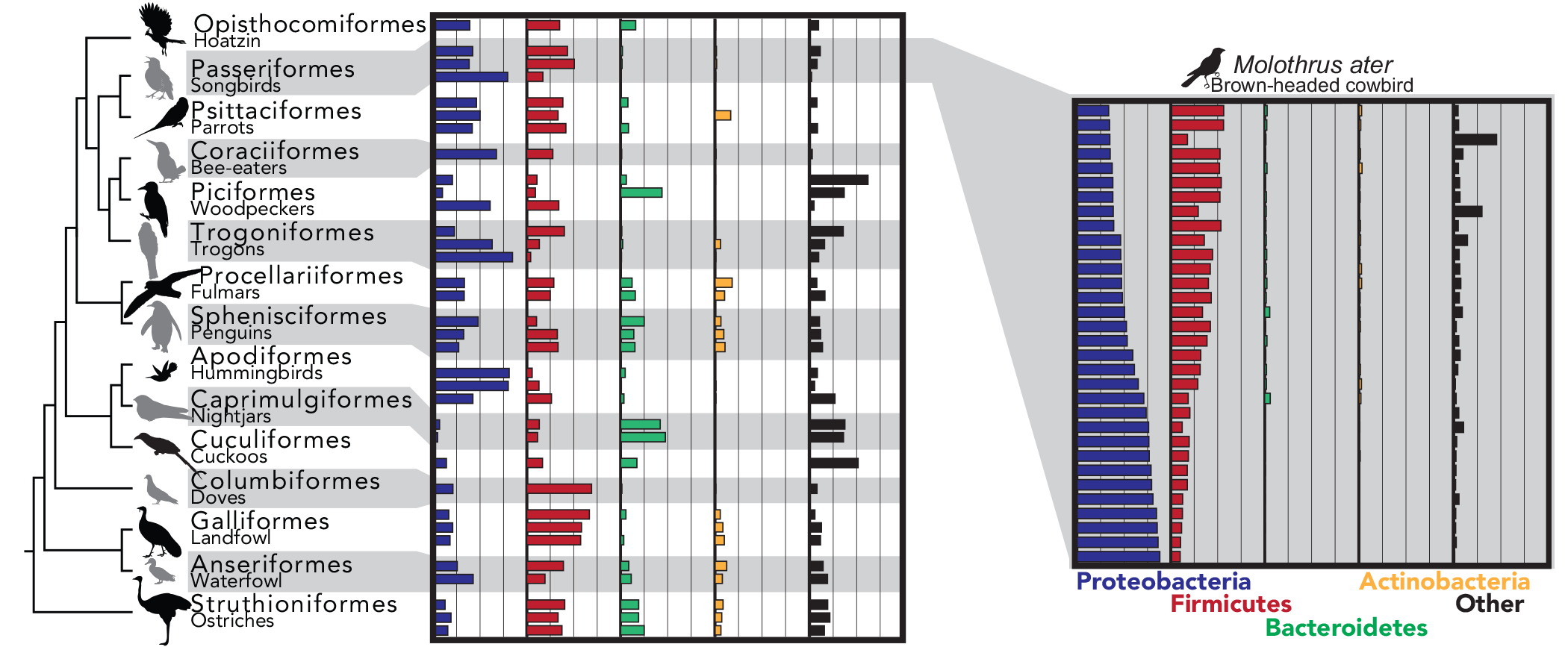
Relative abundance of major bacterial phyla in the gut microbiome of 1-3 individuals from 15 avian orders (left) and from 32 individuals from a single species. Molothrus ater (right). Notice the general patterns (1) Proteobacteria and Fimicutes tend to dominate and (2) there is a large amount of variation between individuals.

Capunitan et al. (2020) cover image for Molecular Ecology.
Bioinformatics
Bioinformatics is a tool we can use to answer biological questions. Many modern biologists benefit from a working knowledge of computers and one or more programming languages. In addition to the practical aspects of bioinformatics, I’m interested in using complex simulation studies to answer questions that cannot be addressed empirically. Finally, I’m always working on effective ways to summarize and display key results from Big Data.
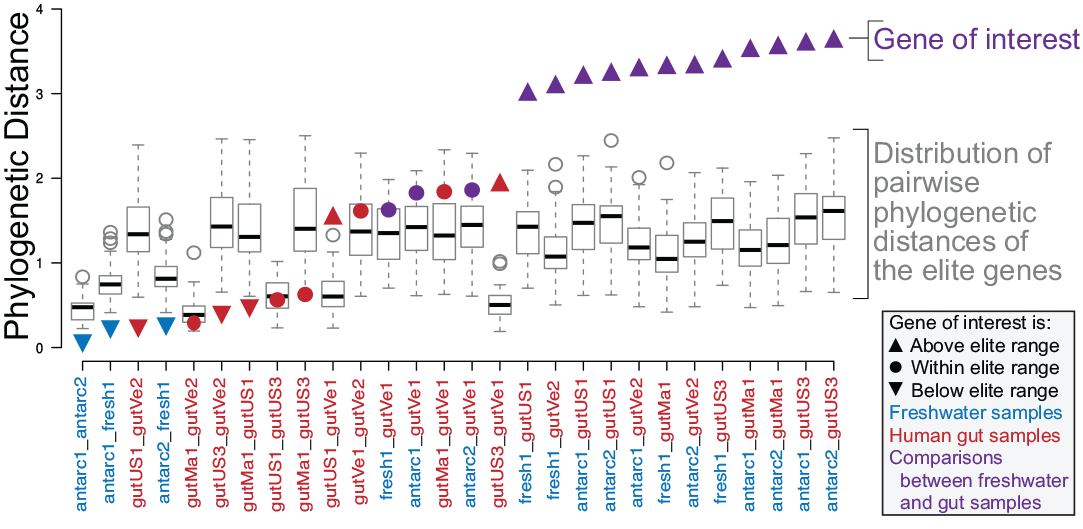
Identifying genes of interest in metagenomes. I’ve written a pipeline that calculates pairwise phylogenetic distances between communities for thousands of gene families. “Elite” genes create a distribution of distances that serve as a specific, pairwise expectation of taxonomic distance. Identifying genes that are outside that expected range of taxonmic diversity indicates genes that may be under unique evolutionary pressures.
Phylogeography
Phylogeography is simply the study of genetic lineages across space and time. Thinking about organisms (big and small) in terms of these dimensions is useful for many aspects of microbiome research and is a framework for thinking about the microbiome. I’m interested in explicitly applying phylogeographic theory to host-associated microbiomes.
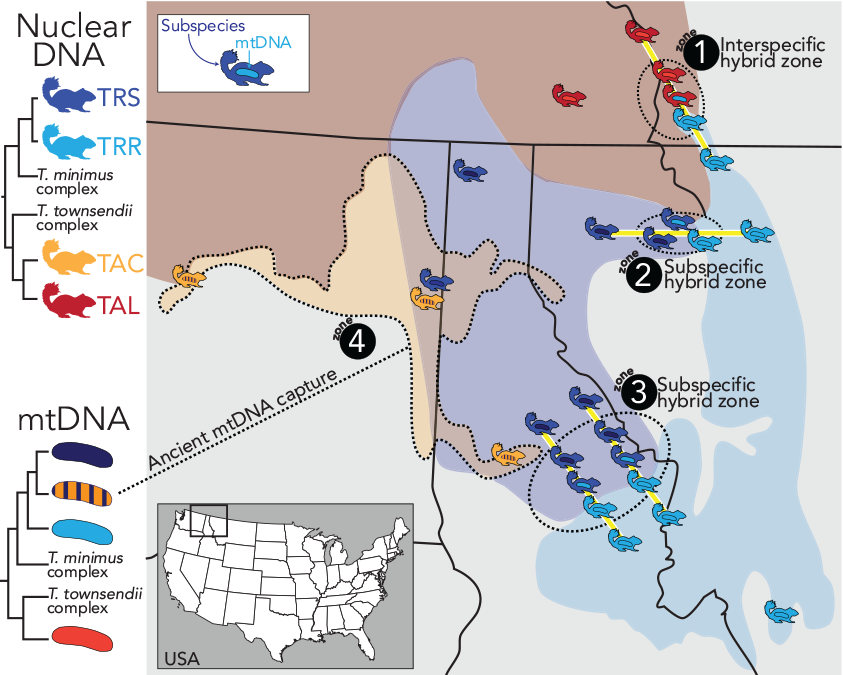
Phylogenetic relationship between chipmunk nuclear genomes (top left) and mitochondrial genomes (bottom left). A map of the mitcohondrial/nuclear types and hybrid zones (1-4) shown at right. Chipmunks also represent proposed sampling localities with transects highlighted in yellow.